PLEASE NOTE: The Phaeodactylum tricornutum genome sequence is composed of "finished chromosomes" (Phatr2) and "unmapped sequence"(Phatr2_bd), which were annotated separately. Please use both portals to make a complete analysis of the genome.
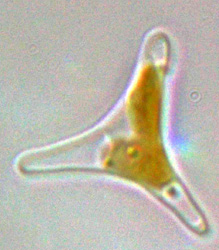
Diatoms are eukaryotic, photosynthetic microorganisms found throughout marine and freshwater ecosystems that are responsible for around 20% of global primary productivity. A defining feature of diatoms is their ornately patterned silicified cell wall (known as frustule), which display species-specific nanoscale-structures. These organisms therefore play major roles in global carbon and silicon cycles.
The marine pennate diatom Phaeodactylum tricornutum is the second diatom for which a whole genome sequence has been generated. It was chosen primarily because of the superior genetic resources available for this diatom (eg, genetic transformation, 100,000 ESTs), and because it has been used in laboratory-based studies of diatom physiology for several decades. Although not considered to be of great ecological significance, it has been found in several locations around the world, typically in coastal areas with wide fluctutations in salinity. Unlike other diatoms it can exist in different morphotypes, and changes in cell shape can be stimulated by environmental conditions. This feature can be used to explore the molecular basis of cell shape control and morphogenesis. Furthermore the species can grow in the absence of silicon, and the biogenesis of silicified frustules is facultative, thereby providing opportunities for experimental exploration of silicon-based nanofabrication in diatoms. The sequence is 30 mega base pairs and, together with the sequence from the centric diatom Thalassiosira pseudonana (34 Mbp; the first diatom whole genome sequence), it provides the basis for comparative genomics studies of diatoms with other eukaryotes and will provide a foundation for interpreting the ecological success of these organisms.
The clone of P. tricornutum that was sequenced is CCAP1055/1 and is available from CCAP. This clone represents a monoclonal culture derived from a fusiform cell in May 2003 from strain CCMP632, which was originally isolated in 1956 off Blackpool (U.K.). It has been maintained in culture continuously in F/2 medium.
The Phaeodactylum genome reveals the evolutionary history of diatom genomes Nature. 2008 Nov 13;456(7219):239-44.